library(tidyverse)
library(tidymodels)
library(rpart) # for building trees
library(rpart.plot) # for plotting trees
library(randomForest) # for bagging & forests
library(infer) # for resampling
library(fivethirtyeight)
data("candy_rankings")15 Random forests & bagging
Settling In
- Sit with the same group as last class.
- Ask your groupmates what classes they are planning to take next semester!
- Locate and open today’s QMD
- Catch up on announcements and messages on Slack
Registration Tips
Thinking about what MSCS course to take next semester? Consider the following:
- Interested in working with data (eg wrangling, plotting, data acquisition, advanced data processing)? Try:
- COMP/STAT 112 (Intro to Data Science)
- COMP/STAT 212 (Intermediate Data Science)
- Interested in data and its connection to public health? Try:
- STAT 125 (Epidemiology)
- STAT 494 (Statistical Genetics)
- Interested in developing a mathematical foundation for better/deeper understanding of statistical methodology? Try:
- MATH/STAT 354 (Probability)
- MATH/STAT 355 (Statistical Theory)
- COMP/MATH 365 (Computational Linear Algebra)
- Interested in learning some computing techniques that are fundamental to data science? Try:
- COMP 123 (Core Concepts in Comp Sci)
- COMP 127 (Object-Oriented Programming and Abstraction)
- COMP 302 (Introduction to Database Management Systems)
NOTE: all of the highlighted courses above require permission of instructor to register. Fill out the corresponding interest form by the end of the day TODAY if you’re interested in taking any of these courses: https://www.macalester.edu/mscs/mscs-course-waitlists/
More generally, use this shiny app (created by Prof Brianna Heggeseth) to help explore courses being offered next semester: https://bheggeseth.shinyapps.io/Spring2025Courses/
Where Are We?
Let’s check out the table of contents in our textbook…
. . .
We’ve covered so far:
- Chapter 2
- Chapter 3
- Chapter 4 (except 4.4–4.6, which we won’t cover)
- Chapter 5
- Chapter 6 (except 6.3, but we’ll get there)
- Chapter 7 (we focused on 7.1, 7.6–7.7)
- Chapter 8 (doing more today…)
. . .
After this, we’ll cover:
- Chapter 12
- Chapter 6.3
. . .
Curious about some of the other chapters?
- Chapters 9 and 13: take Statistical Genetics
- Chapter 10: take Introduction to Artificial Intelligence
- Chapter 11: take Survival Analysis
Small Group Discussion
Discuss the following examples with your group.
Context
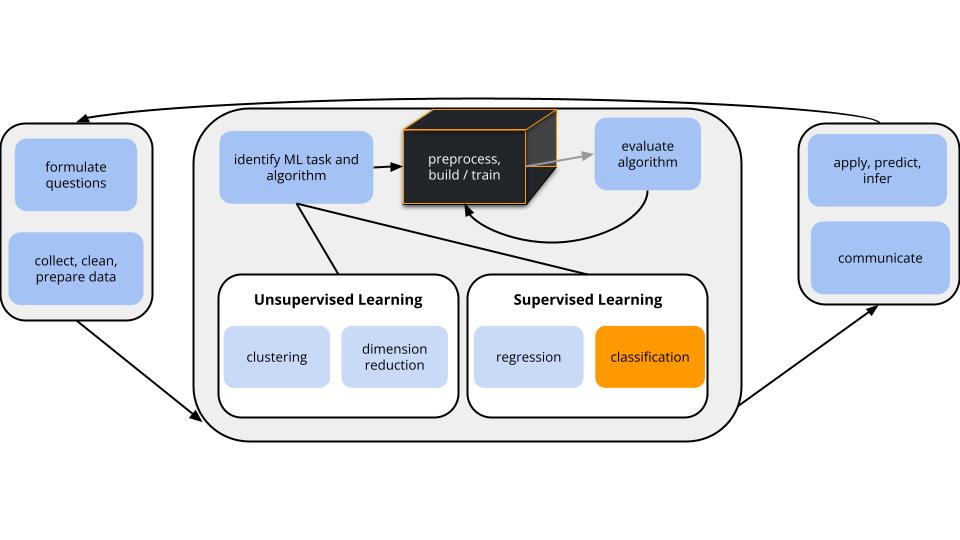
Within the broader machine learning landscape, we left off by discussing supervised classification techniques:
- build a model of categorical variable y by predictors x
- parametric model: logistic regression
- nonparametric models: KNN & trees
- evaluate the model
We can use CV & in-sample techniques to estimate the accuracy of our classification models.- for binary y: sensitivity, specificity, ROC curves
- for y with any number of categories: overall accuracy rates, category specific accuracy rates
TODAY’S GOAL
Add more nonparametric algorithms to our toolkit: random forests & bagging
EXAMPLE 1: Anticipation
What does the word “forest” mean to you?
EXAMPLE 2: Candy!!!
head(candy_rankings)# A tibble: 6 × 13
competitorname chocolate fruity caramel peanutyalmondy nougat crispedricewafer
<chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 100 Grand TRUE FALSE TRUE FALSE FALSE TRUE
2 3 Musketeers TRUE FALSE FALSE FALSE TRUE FALSE
3 One dime FALSE FALSE FALSE FALSE FALSE FALSE
4 One quarter FALSE FALSE FALSE FALSE FALSE FALSE
5 Air Heads FALSE TRUE FALSE FALSE FALSE FALSE
6 Almond Joy TRUE FALSE FALSE TRUE FALSE FALSE
# ℹ 6 more variables: hard <lgl>, bar <lgl>, pluribus <lgl>,
# sugarpercent <dbl>, pricepercent <dbl>, winpercent <dbl>Write R code to find out the following:
# What are the 6 most popular candies?
# The least popular?Solution:
# What are the 6 most popular candies?
## OPTION 1
candy_rankings %>%
arrange(desc(winpercent)) %>%
head()# A tibble: 6 × 13
competitorname chocolate fruity caramel peanutyalmondy nougat crispedricewafer
<chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 Reese's Peanu… TRUE FALSE FALSE TRUE FALSE FALSE
2 Reese's Minia… TRUE FALSE FALSE TRUE FALSE FALSE
3 Twix TRUE FALSE TRUE FALSE FALSE TRUE
4 Kit Kat TRUE FALSE FALSE FALSE FALSE TRUE
5 Snickers TRUE FALSE TRUE TRUE TRUE FALSE
6 Reese's pieces TRUE FALSE FALSE TRUE FALSE FALSE
# ℹ 6 more variables: hard <lgl>, bar <lgl>, pluribus <lgl>,
# sugarpercent <dbl>, pricepercent <dbl>, winpercent <dbl>## OPTION 2
candy_rankings %>%
slice_max(winpercent, n = 6)# A tibble: 6 × 13
competitorname chocolate fruity caramel peanutyalmondy nougat crispedricewafer
<chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 Reese's Peanu… TRUE FALSE FALSE TRUE FALSE FALSE
2 Reese's Minia… TRUE FALSE FALSE TRUE FALSE FALSE
3 Twix TRUE FALSE TRUE FALSE FALSE TRUE
4 Kit Kat TRUE FALSE FALSE FALSE FALSE TRUE
5 Snickers TRUE FALSE TRUE TRUE TRUE FALSE
6 Reese's pieces TRUE FALSE FALSE TRUE FALSE FALSE
# ℹ 6 more variables: hard <lgl>, bar <lgl>, pluribus <lgl>,
# sugarpercent <dbl>, pricepercent <dbl>, winpercent <dbl># The least popular?
## OPTION 1
candy_rankings %>%
arrange(winpercent) %>%
head()# A tibble: 6 × 13
competitorname chocolate fruity caramel peanutyalmondy nougat crispedricewafer
<chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 Nik L Nip FALSE TRUE FALSE FALSE FALSE FALSE
2 Boston Baked … FALSE FALSE FALSE TRUE FALSE FALSE
3 Chiclets FALSE TRUE FALSE FALSE FALSE FALSE
4 Super Bubble FALSE TRUE FALSE FALSE FALSE FALSE
5 Jawbusters FALSE TRUE FALSE FALSE FALSE FALSE
6 Root Beer Bar… FALSE FALSE FALSE FALSE FALSE FALSE
# ℹ 6 more variables: hard <lgl>, bar <lgl>, pluribus <lgl>,
# sugarpercent <dbl>, pricepercent <dbl>, winpercent <dbl>## OPTION 2
candy_rankings %>%
slice_min(winpercent, n = 6)# A tibble: 6 × 13
competitorname chocolate fruity caramel peanutyalmondy nougat crispedricewafer
<chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 Nik L Nip FALSE TRUE FALSE FALSE FALSE FALSE
2 Boston Baked … FALSE FALSE FALSE TRUE FALSE FALSE
3 Chiclets FALSE TRUE FALSE FALSE FALSE FALSE
4 Super Bubble FALSE TRUE FALSE FALSE FALSE FALSE
5 Jawbusters FALSE TRUE FALSE FALSE FALSE FALSE
6 Root Beer Bar… FALSE FALSE FALSE FALSE FALSE FALSE
# ℹ 6 more variables: hard <lgl>, bar <lgl>, pluribus <lgl>,
# sugarpercent <dbl>, pricepercent <dbl>, winpercent <dbl>
EXAMPLE 3: Build an unpruned tree
For demonstration purposes only let’s:
- define a
popularityvariable that categorizes the candies as “low”, “medium”, or “high” popularity - delete the original
winpercentvariable - rename variables to make them easier to read in a tree
- make the candy name a row label, not a predictor
candy <- candy_rankings %>%
mutate(popularity = cut(winpercent, breaks = c(0, 40, 60, 100), labels = c("low", "med", "high"))) %>%
select(-winpercent) %>%
rename("price" = pricepercent, "sugar" = sugarpercent, "nutty" = peanutyalmondy, "wafer" = crispedricewafer) %>%
column_to_rownames("competitorname")Our goal is to model candy popularity by all possible predictors in our data.
# STEP 1: tree specification
tree_spec <- decision_tree() %>%
set_mode("classification") %>%
set_engine(engine = "rpart") %>%
set_args(cost_complexity = 0, min_n = 2, tree_depth = 30)
# STEP 2: Build the tree! No tuning (hence no workflows) necessary.
original_tree <- tree_spec %>%
fit(popularity ~ ., data = candy)
# Plot the tree
original_tree %>%
extract_fit_engine() %>%
plot(margin = 0)
original_tree %>%
extract_fit_engine() %>%
text(cex = 0.7)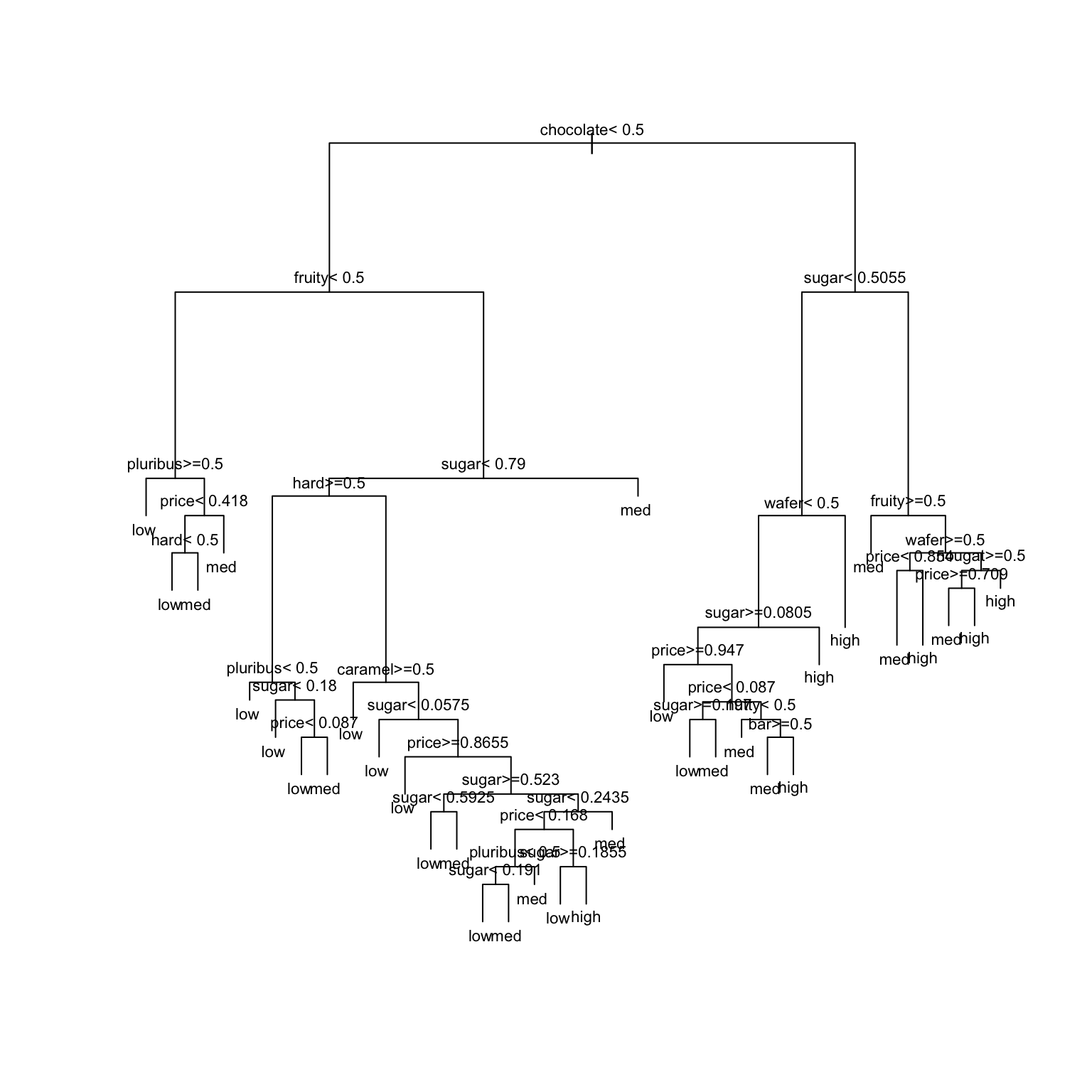
Ideally, our classification algorithm would have both low bias and low variance:
- low variance = the results wouldn’t change much if we changed up the data set
- low bias = within any data set, the predictions of y tend to have low error / high accuracy
Unfortunately, like other overfit algorithms, unpruned trees don’t enjoy both of these. They have…
- low bias, low variance
- low bias, high variance
- high bias, low variance
- high bias, high variance
Solution:
low bias, high variance
New Concept
GOAL
Maintain the low bias of an unpruned tree while decreasing variance.
APPROACH
Build a bunch of unpruned trees from different data. This way, our final result isn’t overfit to our sample data.
THE RUB (CHALLENGE/DIFFICULTY)
We only have 1 set of data…
EXAMPLE 4: Take a REsample of candy
We only have 1 sample of data. But we can resample it (basically pretending we have a different sample).
Let’s each take our own unique candy resample (aka bootstrapping):
- Take a sample of 85 candies from the original 85 candies, with replacement.
- Some data points will be sampled multiple times while others aren’t sampled at all.
- On average, 2/3 of the original data points will show up in the resample and 1/3 will be left out.
Take your resample:
# Set the seed to YOUR phone number (just the numbers)
set.seed(123456789)
# Take a REsample of candies from our sample
my_candy <- sample_n(candy, size = nrow(candy), replace = TRUE)
# Check it out
head(my_candy, 3) chocolate fruity caramel nutty nougat wafer hard bar
Snickers Crisper...1 TRUE FALSE TRUE TRUE FALSE TRUE FALSE TRUE
Fruit Chews...2 FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
Nestle Crunch...3 TRUE FALSE FALSE FALSE FALSE TRUE FALSE TRUE
pluribus sugar price popularity
Snickers Crisper...1 FALSE 0.604 0.651 med
Fruit Chews...2 TRUE 0.127 0.034 med
Nestle Crunch...3 FALSE 0.313 0.767 highIn the next exercise, we’ll each build a tree of popularity using our own resample data.
First, check your intuition:
- TRUE / FALSE: All of our trees will be the same.
- TRUE / FALSE: Our trees will use the same predictor (but possibly a different cut-off) in the first split.
- TRUE / FALSE: Our trees will use the same predictors in all splits.
Solution:
- FALSE
- FALSE
- FALSE
Fun Math Facts:
With resampling (also known as bootstrapping), we have an original sample of \(n\) rows. We drawn individual rows with replacement from this set until we have another set of size \(n\).
The probability of choosing any one row (say the 1st row) on the first draw is \(1/n\). The probability of not choosing that one row is \(1-1/n\). That is just for the first draw. There are \(n\) draws, all of which are independent, so the probability of never choosing this particular row on any of the draws is \((1-1/n)^n\).
If we consider larger and larger datasets (large \(n\) going to infinity), then
\[\lim_{n \rightarrow \infty} (1-1/n)^n = 1/e \approx 0.368\]
Thus, the probability that any one row is NOT chosen is about 1/3 and the probability that any one row is chosen is 2/3.
EXAMPLE 6: Using our FOREST
We now have a group of multiple trees – a forest!
These trees…
- differ from resample to resample
- don’t use the same predictor in each split (not even in the first split)!
- produce different
popularitypredictions for Baby Ruth
Based on our forest of trees (not just your 1 tree), what’s your prediction for Baby Ruth’s popularity?
What do you think are the advantages of predicting candy popularity using a forest instead of a single tree?
Can you anticipate any drawbacks of using forests instead of trees?
Solution:
- take the majority vote, i.e. most common category
- by averaging across multiple trees, classifications will be more stable / less variable from dataset to dataset (lower variance)
- computational intensity (lack of efficiency)
Notes: Bagging and Forests
BAGGING (Bootstrap AGGregatING) & Random Forests
To classify a categorical response variable y using a set of p predictors x:
Take B resamples from the original sample.
- Sample WITH replacement
- Sample size = original sample size n
Use each resample to build an unpruned tree.
- For bagging: consider all p predictors in each split of each tree
- For random forests: at each split in each tree, randomly select and consider only a subset of the predictors (often roughly p/2 or \(\sqrt{p}\))
- For bagging: consider all p predictors in each split of each tree
Use each of the B trees to classify y at a set of predictor values x.
Average the classifications using a majority vote: classify y as the most common classification among the B trees.
Ensemble Methods
Bagging and random forest algorithms are ensemble methods.
They combine the outputs of multiple machine learning algorithms.
As a result, they decrease variability from sample to sample, hence provide more stable predictions / classifications than might be obtained by any algorithm alone.
EXAMPLE 7: pros & cons
- Order trees, forests, & bagging algorithms from least to most computationally expensive.
- What results will be easier to interpret: trees or forests?
- Which of bagging or forests will produce a collection of trees that tend to look very similar to each other, and similar to the original tree? Hence which of these algorithms is more dependent on the sample data, thus will vary more if we change up the data? [both questions have the same answer]
Solution:
- trees, forests, bagging
- trees (we can’t draw a forest)
- bagging (forests tend to have lower variability)
Exercises
For the rest of the class, work together on Exercises 1–7
- Tuning parameters (challenge)
Our random forest of popularity by all 11 possible predictors will depend upon 3 tuning parameters:
trees= the number of trees in the forestmtry= number of predictors to randomly choose & consider at each splitmin_n= minimum number of data points in any leaf node of any tree
Check your intuition.
- Does increasing the number of
treesmake the forest algorithm more or less variable from dataset to dataset? - We have 11 possible predictors, and sqrt(11) is roughly 3. Recall: Would considering just 3 randomly chosen predictors in each split (instead of all 11) make the forest algorithm more or less variable from dataset to dataset?
- Recall that using unpruned trees in our forest is important to maintaining low bias. Thus should
min_nbe small or big?
Solution:
- less variable (less impacted by “unlucky” trees)
- less variable
- small
- Build the forest
Given that forests are relatively computationally expensive, we’ll only build one forest using the following tuning parameters:
mtry = NULL: this setsmtryto the default, which is sqrt(number of predictors)trees = 500min_n = 2
Fill in the below code to run this forest algorithm.
# There's randomness behind the splits!
set.seed(253)
# STEP 1: Model Specification
rf_spec <- rand_forest() %>%
set_mode("___") %>%
___(engine = "ranger") %>%
___(
mtry = NULL,
trees = 500,
min_n = 2,
probability = FALSE, # Report classifications, not probability calculations
importance = "impurity" # Use Gini index to measure variable importance
)
# STEP 2: Build the forest
# There are no preprocessing steps or tuning, hence no need for a workflow!
candy_forest <- ___ %>%
fit(___, ___)Solution:
# There's randomness behind the splits!
set.seed(253)
# STEP 1: Model Specification
rf_spec <- rand_forest() %>%
set_mode("classification") %>%
set_engine(engine = "ranger") %>%
set_args(
mtry = NULL,
trees = 500,
min_n = 2,
probability = FALSE, # give classifications, not probability calculations
importance = "impurity" # use Gini index to measure variable importance
)
# STEP 2: Build the forest
# There are no preprocessing steps or tuning, hence no need for a workflow!
candy_forest <- rf_spec %>%
fit(popularity ~ ., data = candy)- Use the forest for prediction
Use the forest to predict thepopularitylevel for Baby Ruth. (Remember that its realpopularityis “med”.)
candy_forest %>%
predict(new_data = candy[7,])# A tibble: 1 × 1
.pred_class
<fct>
1 med - Evaluating forests: concepts
But how good is our forest at classifying candy popularity?
To this end, we could evaluate 3 types of forest predictions.
- Why don’t in-sample predictions, i.e. asking how well our forest classifies our sample candies, give us an “honest” assessment of our forest’s performance?
- Instead, suppose we used 10-fold cross-validation (CV) to estimate how well our forest classifies new candies. In this process, how many total trees would we need to construct?
- Alternatively, we can estimate how well our forest classifies new candies using the out-of-bag (OOB) error rate. Since we only use a resample of data points to build any given tree in the forest, the “out-of-bag” data points that do not appear in a tree’s resample are natural test cases for that tree. The OOB error rate tracks the proportion or percent of these out-of-bag test cases that are misclassified by their tree. How many total trees would we need to construct to calculate the OOB error rate?
- Moving forward, we’ll use OOB and not CV to evaluate forest performance. Why?
Solution:
- they use the same data we used to build the forest
- 10 forests
*500 trees each = 5000 trees - 1 forest
*500 trees = 500 trees - it’s much more computationally efficient
- Evaluating forests: implementation
- Report and interpret the estimated
OOB prediction error.
candy_forestparsnip model object
Ranger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, num.trees = ~500, min.node.size = min_rows(~2, x), probability = ~FALSE, importance = ~"impurity", num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1))
Type: Classification
Number of trees: 500
Sample size: 85
Number of independent variables: 11
Mtry: 3
Target node size: 2
Variable importance mode: impurity
Splitrule: gini
OOB prediction error: 40.00 % - The test or OOB confusion matrix provides more detail. Use this to confirm the OOB prediction error from part a. HINT: Remember to calculate error (1 - accuracy), not accuracy.
# NOTE: t() transposes the confusion matrix so that
# the columns and rows are in the usual order
candy_forest %>%
extract_fit_engine() %>%
pluck("confusion.matrix") %>%
t() true
predicted low med high
low 8 6 1
med 15 29 6
high 2 4 14Which level of candy popularity was least accurately classified by our forest?
Check out the in-sample confusion matrix. In general, are the in-sample predictions better or worse than the OOB predictions?
# The cbind() includes the original candy data
# alongside their predicted popularity levels
candy_forest %>%
predict(new_data = candy) %>%
cbind(candy) %>%
conf_mat(
truth = popularity,
estimate = .pred_class
) Truth
Prediction low med high
low 18 0 0
med 6 39 2
high 1 0 19Solution:
- We expect our forest to misclassify roughly 40% of new candies.
- .
# APPROACH 1: # of MISclassifications / total # of classifications
(6 + 1 + 15 + 6 + 2 + 4) / (8 + 29 + 14 + 6 + 1 + 15 + 6 + 2 + 4) [1] 0.4# APPROACH 2: overall MISclassification rate = 1 - overall accuracy rate
# overall accuracy rate
(8 + 29 + 14) / (8 + 29 + 14 + 6 + 1 + 15 + 6 + 2 + 4) [1] 0.6# overall misclassification rate
1 - 0.6[1] 0.4- low (more were classified as “med” than as “low”)
- much better!
- Variable importance
Variable importance metrics, averaged over all trees, measure the strength of the 11 predictors in classifying candy popularity:
# Print the metrics
candy_forest %>%
extract_fit_engine() %>%
pluck("variable.importance") %>%
sort(decreasing = TRUE) sugar price chocolate fruity nutty wafer pluribus bar
9.4075750 9.0830343 5.0757837 2.8236876 2.2084982 2.1255274 1.8596475 1.8143452
caramel hard nougat
1.6918086 1.3640697 0.9441936 # Plot the metrics
library(vip)
candy_forest %>%
vip(geom = "point", num_features = 11)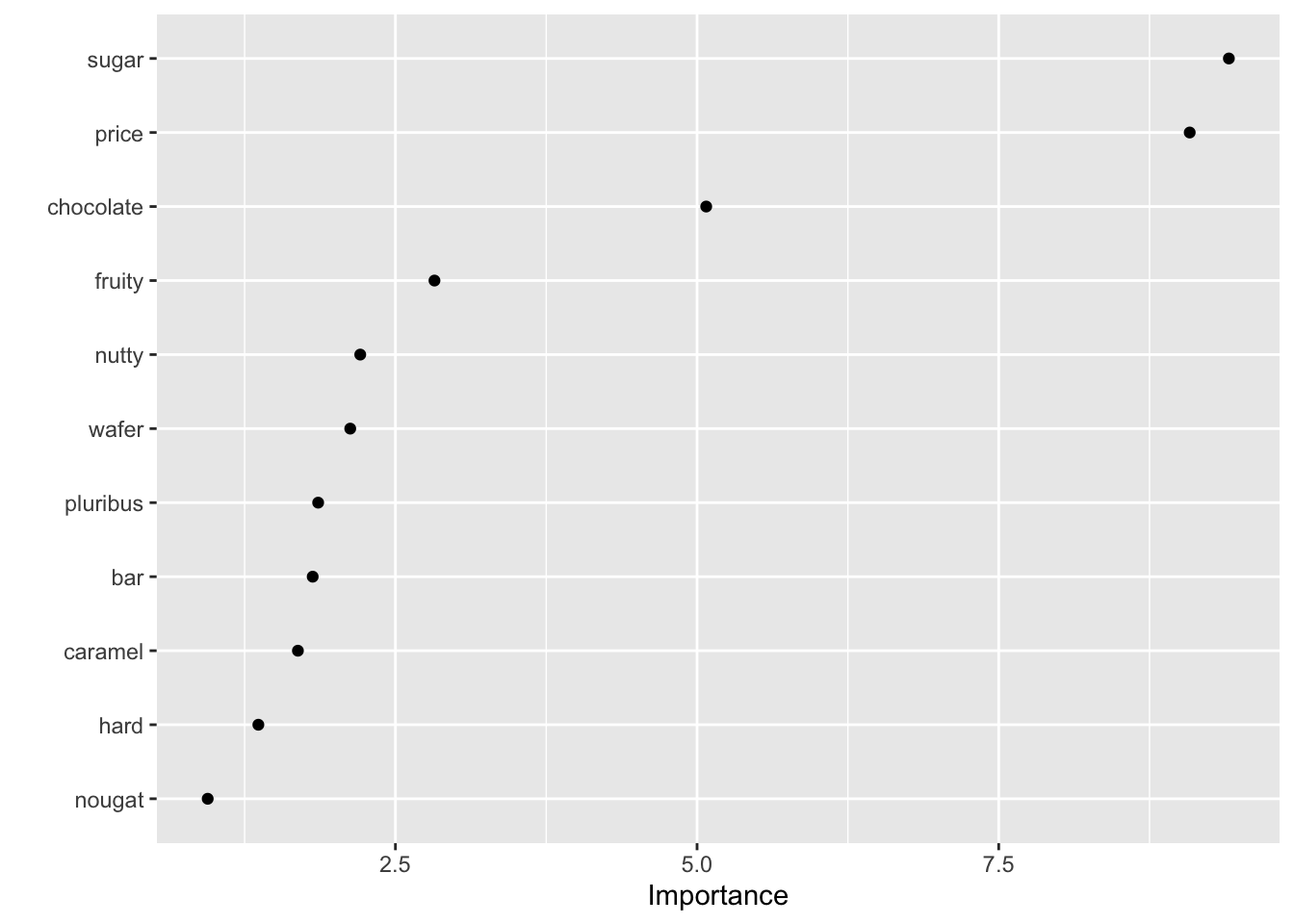
- If you’re a candy connoisseur, does this ranking make some contextual sense to you?
- The only 2 quantitative predictors,
sugarandprice, have the highest importance metrics. This could simply be due to their quantitative structure: trees tend to favor predictors with lots of unique values. Explain. HINT: A tree’s binary splits are identified by considering every possible cut / split point in every possible predictor.
Solution:
- will vary
- predictors with lots of unique values have far more possible split points to choose from
- Classification regions
Just like any classification model, forests divide our data points into classification regions.
Let’s explore this idea using some simulated data that illustrate some important contrasts.1
Import and plot the data:
# Import data
simulated_data <- read.csv("https://kegrinde.github.io/stat253_coursenotes/data/circle_sim.csv") %>%
mutate(class = as.factor(class))
# Plot data
ggplot(simulated_data, aes(y = X2, x = X1, color = class)) +
geom_point() +
theme_minimal()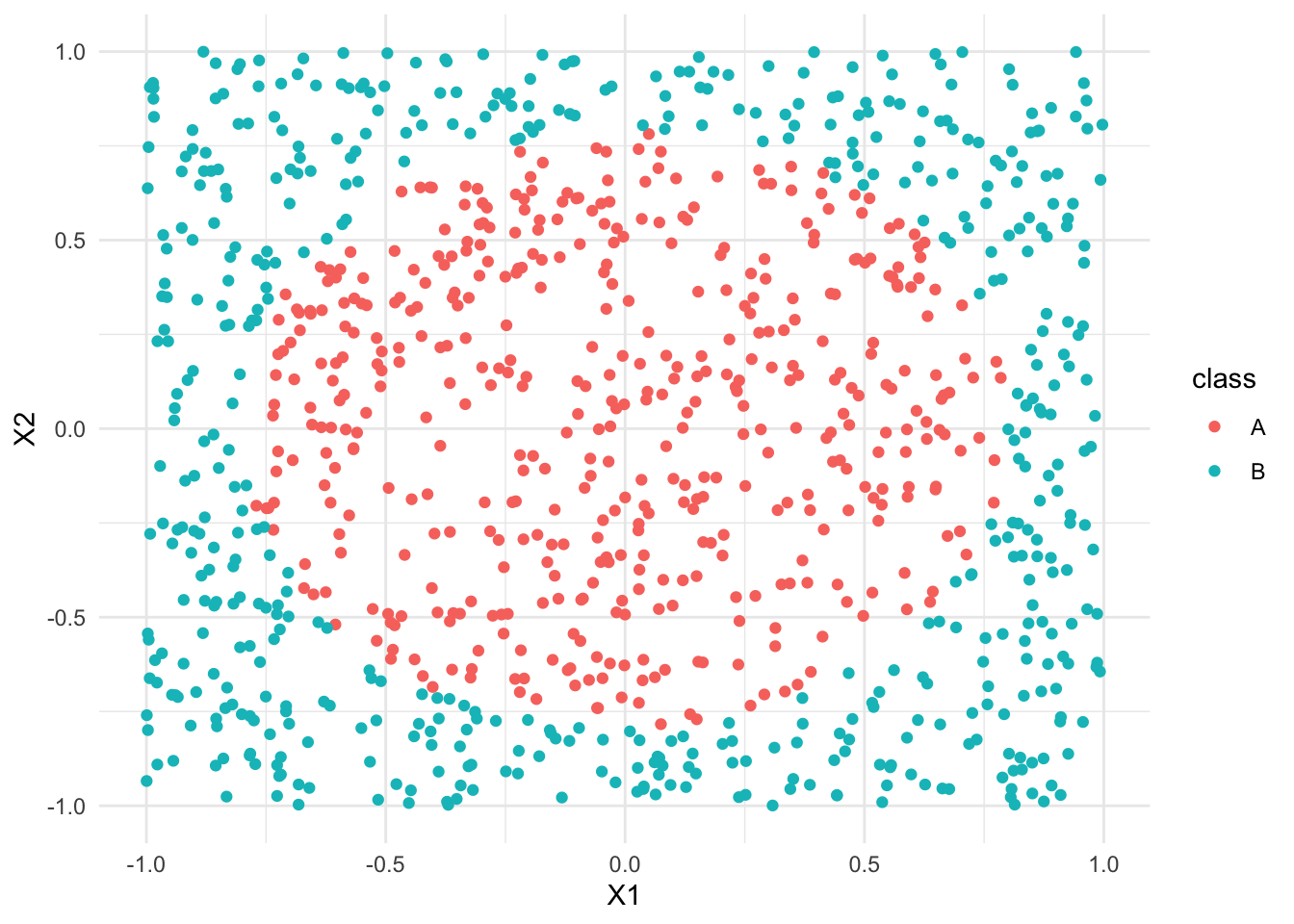
- Below is a classification tree of
classbyX1andX2. What do you think its classification regions will look like?
# Build the (default) tree
circle_tree <- decision_tree() %>%
set_mode("classification") %>%
set_engine(engine = "rpart") %>%
fit(class ~ ., data = simulated_data)
circle_tree %>%
extract_fit_engine() %>%
rpart.plot()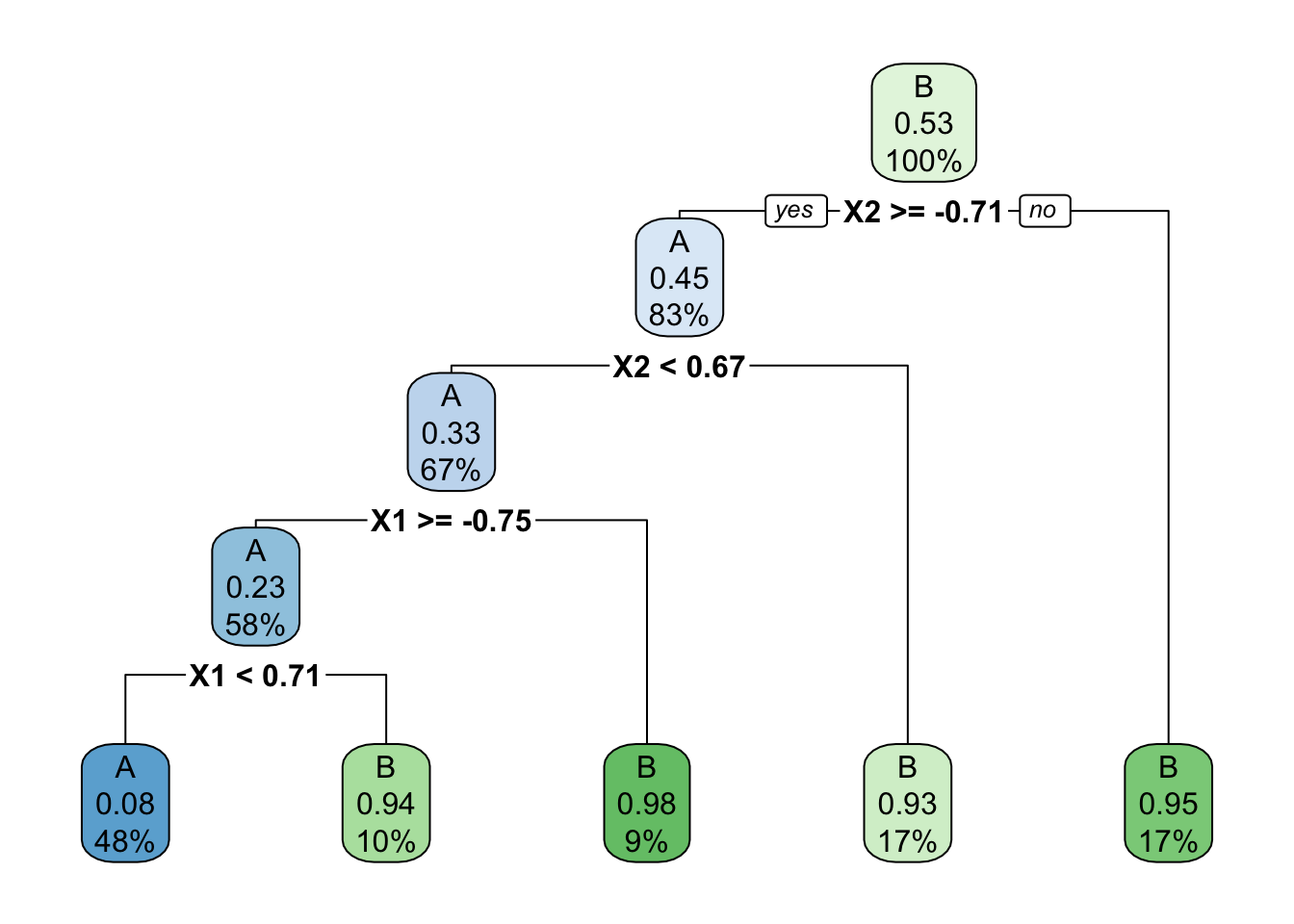
- Check your intuition. Were you right?
# THIS IS ONLY DEMO CODE.
# Plot the tree classification regions
examples <- data.frame(X1 = seq(-1, 1, len = 100), X2 = seq(-1, 1, len = 100)) %>%
expand.grid()
circle_tree %>%
predict(new_data = examples) %>%
cbind(examples) %>%
ggplot(aes(y = X2, x = X1, color = .pred_class)) +
geom_point() +
labs(title = "tree classification regions") +
theme_minimal()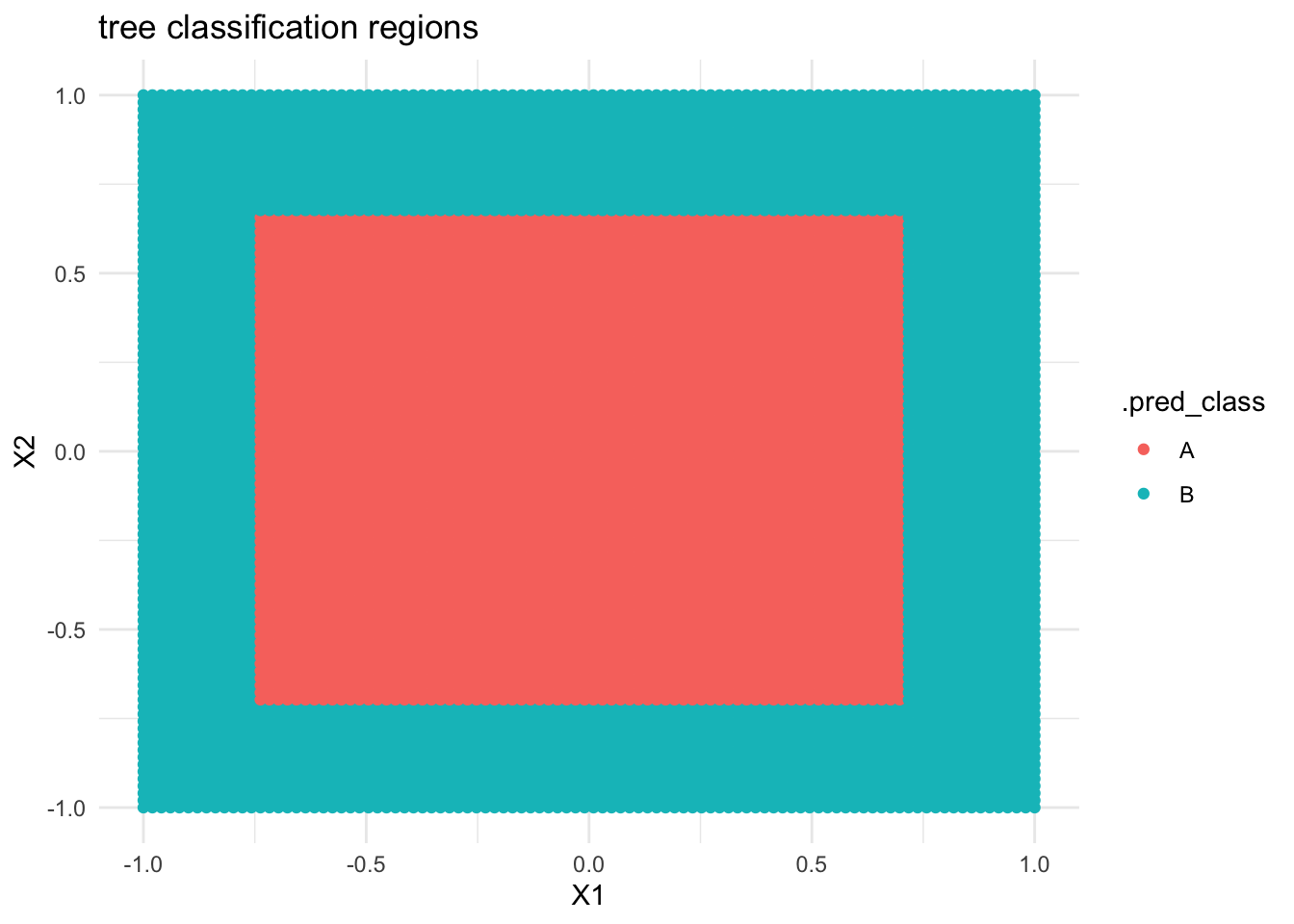
If we built a forest model of
classbyX1andX2, what do you think the classification regions will look like?Check your intuition. Were you right?
# THIS IS ONLY DEMO CODE.
# Build the forest
circle_forest <- rf_spec %>%
fit(class ~ ., data = simulated_data)
# Plot the tree classification regions
circle_forest %>%
predict(new_data = examples) %>%
cbind(examples) %>%
ggplot(aes(y = X2, x = X1, color = .pred_class)) +
geom_point() +
labs(title = "forest classification regions") +
theme_minimal()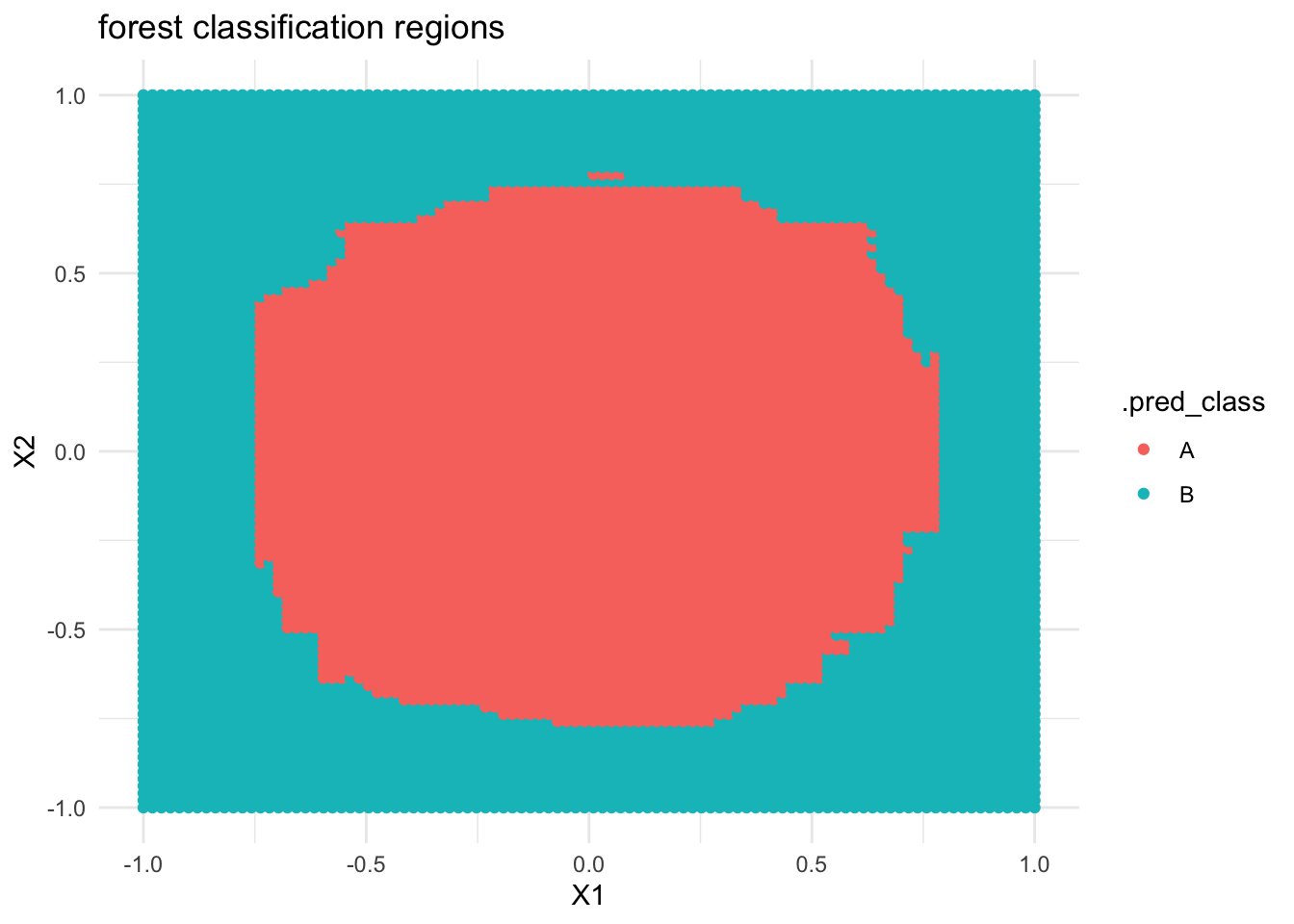
- Reflect on what you’ve observed here!
Solution:
- …
# THIS IS ONLY DEMO CODE.
# Plot the tree classification regions
examples <- data.frame(X1 = seq(-1, 1, len = 100), X2 = seq(-1, 1, len = 100)) %>%
expand.grid()
circle_tree %>%
predict(new_data = examples) %>%
cbind(examples) %>%
ggplot(aes(y = X2, x = X1, color = .pred_class)) +
geom_point() +
labs(title = "tree classification regions") +
theme_minimal()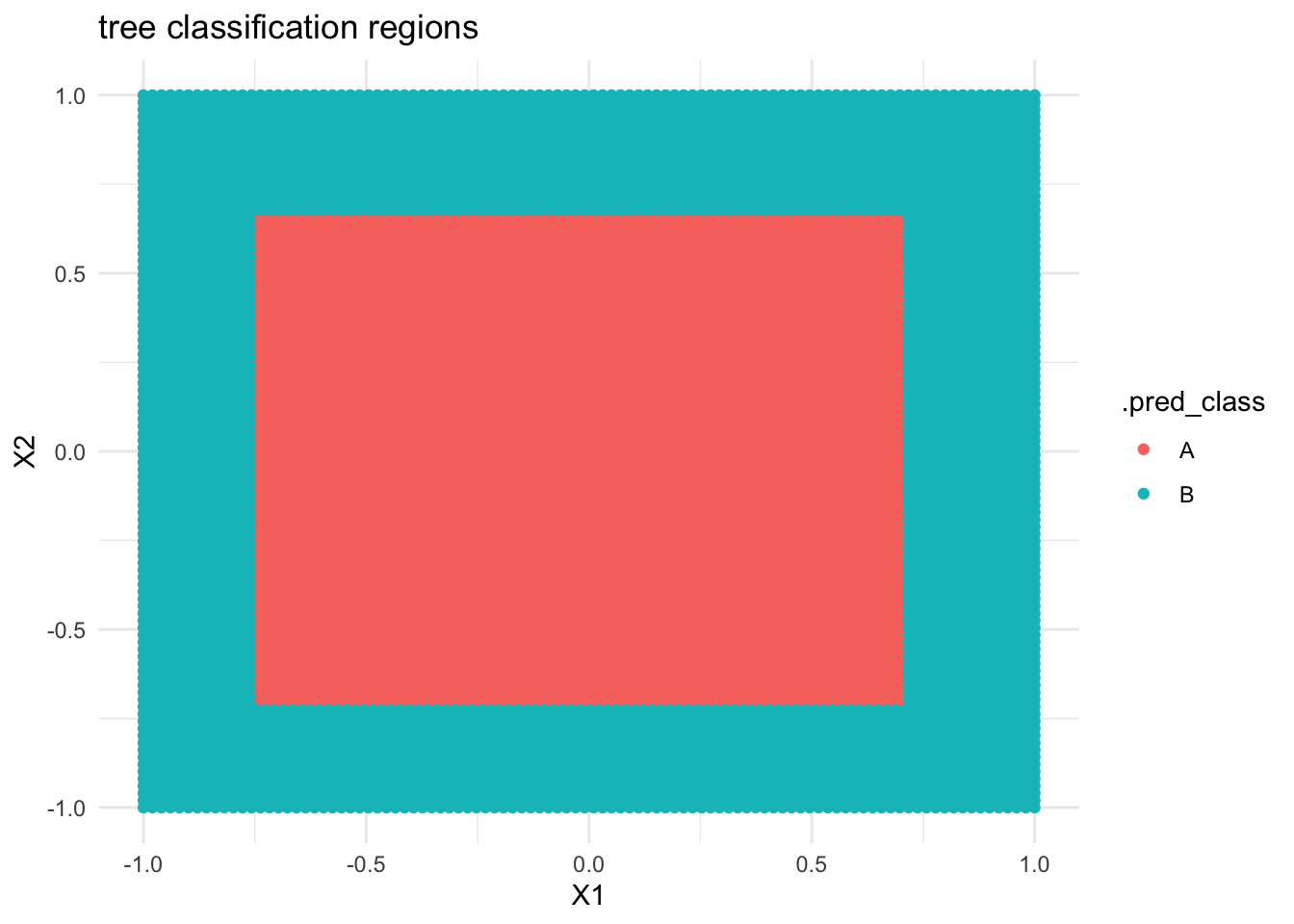
- …
# THIS IS ONLY DEMO CODE.
# Build the forest
circle_forest <- rf_spec %>%
fit(class ~ ., data = simulated_data)
# Plot the tree classification regions
circle_forest %>%
predict(new_data = examples) %>%
cbind(examples) %>%
ggplot(aes(y = X2, x = X1, color = .pred_class)) +
geom_point() +
labs(title = "forest classification regions") +
theme_minimal()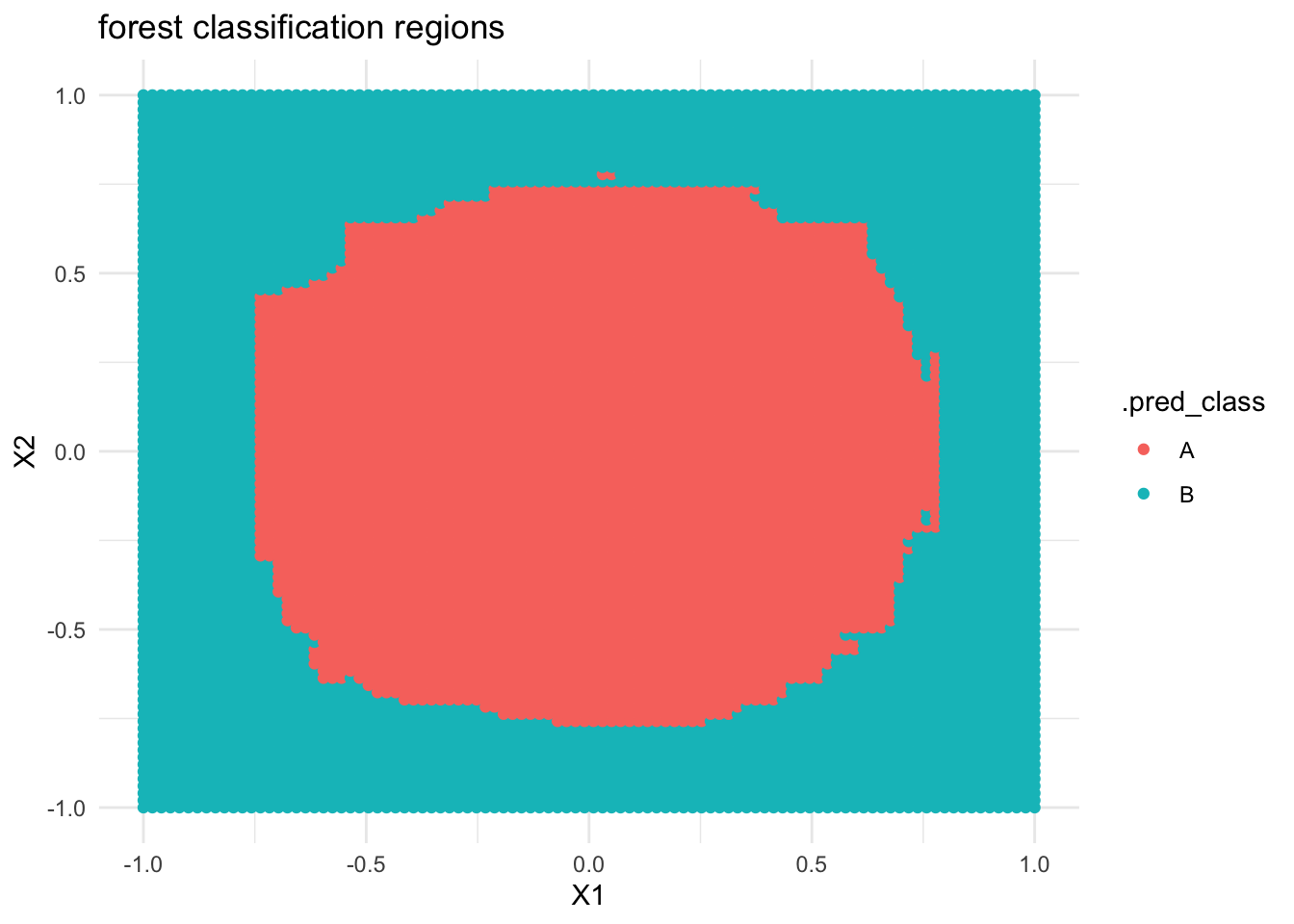
- Forest classification regions are less rigid / boxy than tree classification regions.
If you finish early
Do one of the following:
- Check out the optional “Deeper learning” section below on another ensemble method: boosting.
- Check out group assignment 2 on Moodle. Next class, your group will pick what topic to explore.
- Work on homework.
Wrapping Up
- As usual, take time after class to finish any remaining exercises, check solutions, reflect on key concepts from today, and come to office hours with questions
- Upcoming due dates:
- Before next class: CP11 (formal review of forests) AND review Group Assignment 2 instructions
- Next Wednesday: HW5 and HW4 Revisions
- Coming soon: Quiz 2 (Nov 19), Group Assignment 2 (Nov 26)
Deeper learning (optional)
Extreme gradient boosting, or XGBoost, is yet another ensemble algorithm for regression and classification. We’ll consider the big picture here. If you want to dig deeper:
- Section 8.2.3 of the book provides a more detailed background
- Julia Silge’s blogpost on predicting home runs provides an example of implementing XGBoost using
tidymodels.
The big picture:
Like bagging and forests, boosting combines predictions from B different trees.
BUT these trees aren’t built from B different resamples. Boosting trees are grown sequentially, each tree slowly learning from the previous trees in the sequence to improve in areas where the previous trees didn’t do well. Loosely speaking, data points with larger misclassification rates among previous trees are given more weight in building future trees.
Unlike in bagging and forests, trees with better performance are given more weight in making future classifications.
Bagging vs boosting
Bagging typically helps decrease variance, but not bias. Thus it is useful in scenarios where other algorithms are unstable and overfit to the sample data.
Boosting typically helps decrease bias, but not variance. Thus it is useful in scenarios where other algorithms are stable, but overly simple.
Notes: R code
Suppose we want to build a forest or bagging algorithm of some categorical response variable y using predictors x1 and x2 in our sample_data.
# Load packages
library(tidymodels)
library(rpart)
library(rpart.plot)
# Resolves package conflicts by preferring tidymodels functions
tidymodels_prefer()
Make sure that y is a factor variable
sample_data <- sample_data %>%
mutate(y = as.factor(y))
Build the forest / bagging model
We’ll typically use the following tuning parameters:
trees= 500 (the more trees we use, the less variable the forest)min_n= 2 (the smaller we allow the leaf nodes to be, the less pruned, hence less biased our forest will be)mtry- for forests:
mtry = NULL(the default) will use the “floor”, or biggest integer below, sqrt(number of predictors) - for bagging: set
mtryto the number of predictors
- for forests:
# STEP 1: Model Specification
rf_spec <- rand_forest() %>%
set_mode("classification") %>%
set_engine(engine = "ranger") %>%
set_args(
mtry = ___,
trees = 500,
min_n = 2,
probability = FALSE, # give classifications, not probability calculations
importance = "impurity" # use Gini index to measure variable importance
)
# STEP 2: Build the forest or bagging model
# There are no preprocessing steps or tuning, hence no need for a workflow!
ensemble_model <- rf_spec %>%
fit(y ~ x1 + x2, data = sample_data)
Use the model to make predictions / classifications
# Put in a data.frame object with x1 and x2 values (at minimum)
ensemble_model %>%
predict(new_data = ___)
Examine variable importance
# Print the metrics
ensemble_model %>%
extract_fit_engine() %>%
pluck("variable.importance") %>%
sort(decreasing = TRUE)
# Plot the metrics
# Plug in the number of top predictors you wish to plot
# (The upper limit varies by application!)
library(vip)
ensemble_model %>%
vip(geom = "point", num_features = ___)
Evaluate the classifications
# Out-of-bag (OOB) prediction error
ensemble_model
# OOB confusion matrix
ensemble_model %>%
extract_fit_engine() %>%
pluck("confusion.matrix") %>%
t()
# In-sample confusion matrix
ensemble_model %>%
predict(new_data = sample_data) %>%
cbind(sample_data) %>%
conf_mat(
truth = y,
estimate = .pred_class
)citation: https://daviddalpiaz.github.io/r4sl/ensemble-methods.html#tree-versus-ensemble-boundaries↩︎